CellAgent
An LLM-driven multi-agent framework for single-cell data analysis,
ensuring high-quality results with minimal effort.
An LLM-driven multi-agent framework for single-cell data analysis,
ensuring high-quality results with minimal effort.
1. Quality Control
2. Normalization
3. Identification of Highly Variable Genes
4. Dimensionality Reduction
5. Clustering
6. Cell Type Annotation# Filter based on the properties of current dataset and empirical thresholds
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=3)
adata.var['mt'] = adata.var_names.str.startswith('MT-')
# Calculate quality control metrics
sc.pp.calculate_qc_metrics(adata, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
# Filter cells with less than 2500 genes and mitochondrial gene percentage less than 5%
adata = adata[adata.obs.n_genes_by_counts < 2500, :]
adata = adata[adata.obs.pct_counts_mt < 5, :]
# Other steps omitted
sc.tl.pca(adata, svd_solver='arpack')
# For this dataset, n_neighbors=10 and n_pcs=40 show clear results
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
sc.tl.leiden(adata)
sc.tl.umap(adata)
sc.pl.umap(adata, color=['leiden']) "Based on the current data analysis results, and in biomedical research, these genes can typically serve as markers to help us identify different cell types..."
"Based on the current data analysis results, and in biomedical research, these genes can typically serve as markers to help us identify different cell types..." # Find differentially expressed genes for each cluster
sc.tl.rank_genes_groups(adata, 'leiden', method='t-test')
sc.pl.rank_genes_groups(adata, n_genes=20, sharey=False)
#After comparing the differentially expressed genes in each cluster, and under the guidance of experts, cell type labels were assigned to each cluster.
cluster_annotations = {
'0': 'T cells','1': 'B cells','2': 'NK cells','3': 'Monocytes','4': 'Dendritic cells'...
}
adata.obs['cell_type'] = adata.obs['leiden'].map(cluster_annotations)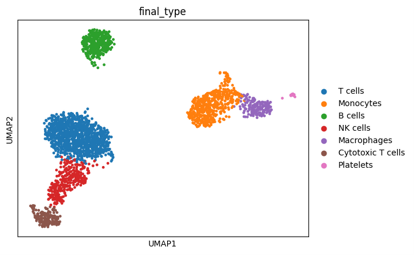
* In typical single-cell data analysis tasks, CellAgent's performance can reach 107.23% compared to the widely used and effective existing algorithms.
** The tasks mentioned here primarily include batch correction, cell type annotation, trajectory inference, spatial domain identification, and spatial transcriptomics imputation, corresponding to existing algorithms such as Scanorama, CellMarker, Slingshot, DeepST, and Tangram, respectively.
CellAgent can streamline your single-cell data analysis workflow, allowing you to obtain high-quality results without the need for complex coding. Whether you are a domain expert or a novice, our online platform enables effortless data analysing and interpretation. With CellAgent, you can:
Considering the double-blind review principle, all external links on this page are hidden during the paper submission period, and this part of the description is temporarily invisible.